
LABTOO Biotech Solutions
Biological samples and experimental models for R&D
Experience the cutting-edge contract research service for biospecimen and experimental outsourcing.
Tailor your own request and create a personalized project with our bespoke services, designed to meet your specific needs.
Begin your journey with an exciting initial project, or explore the exclusive possibilities for your challenge with our premium options.
The first scientific outsourcing management solution
With the complexity of biology comes the complexity of your projects. It is essential to have access to the resources of various laboratories and hospitals.
Understanding the challenges of Biotech
The Labtoo method allows for finding solutions quickly at all stages of the R&D process and in any type of therapeutic area. The scientific team takes time to study and comprehend your projects and their challenges to identify the best solution possible.
For unique R&D requirements, tailor-made solutions
Our team is aware of the complexity behind each R&D project, and our projects are always unique. Adapting to singularity is part of our DNA and a reflection of our success.

Biological Samples Sourcing
Our methodology is well adapted to finding sources of human biological samples like hospitals. We have an exceptional track record of finding rare and unique samples for our clients.
Experimental Services Outsourcing
The Labtoo method was developed around the outsourcing support for preclinical R&D: in vitro models, in vivo models, genomics, and proteomics services. When it is difficult to find, our team is here to support you.


COVID-19 Research
We have adapted our tools and method to provide the best solutions for experimentation on SARS-CoV-2 and other viruses. Find animal models, in vitro models, and diagnostic validation tools to get your project started.
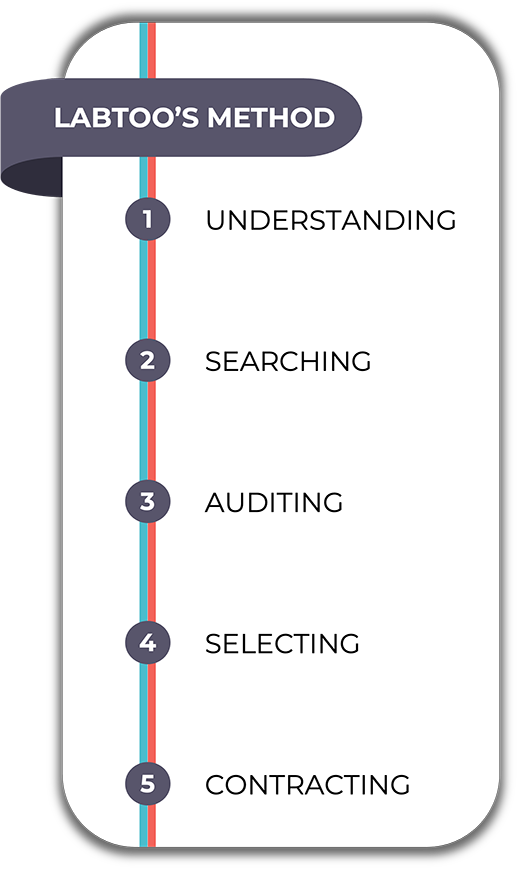
A successful outsourcing
Fast, simple, and secure, at last!
In order to respond to any type of unique demand in the R&D pathway, we need to invent new ways of providing services in biotechnologies and pharma. This is what we have been proposing to innovative project leaders since the creation of our solution.
Whether you are the founder of a biotech startup company, an academic research director, or a project manager at a pharmaceutical company, you regularly need to look for external service providers.
However, the process of R&D outsourcing is very complex. From the identification of technological solutions to contractualization with trusted partners, additional support can significantly increase productivity. For the project leader: security and significant time savings.