Proteomics Services for Drug Development
Proteomics is the study of all the proteins of an organism, an organ, a cell, and even a cell compartment. Proteins are the direct result of genome expression, so two organisms of totally different appearances can have a very similar genome. It is proteins that differentiate them and give them specific characteristics.
Proteomics answers questions about proteins at several levels:
- What proteins are present in a sample?
Millions of different proteins exist in each cell, resulting from genome translation and post-translational modifications: phosphorylation, truncation, ubiquitination, etc. - How many proteins of interest does a sample contain?
Some proteins may be very abundant in a cell, while others will be difficult to detect. - What is the structure of the protein or complex?
There are four levels of protein structure: amino acid succession (primary), alpha-helix and beta-helix fundamental structural elements (secondary), three-dimensional folding using covalent or non-covalent bonds (tertiary), and integration of the protein into a complex (quaternary). - What is the biological state of the protein of interest?
Some proteins undergo profound modifications during their existence: phosphorylation and dephosphorylation, truncations, degradation, etc.
Proteomics services cover a wide spectrum of analyses of proteins and peptides present in a sample. For example, it is about identifying proteins by separating a mixture by liquid chromatography followed by detection by tandem mass spectrometry (LC-MS/MS).
New techniques of cell separation and improved performance of proteomics instruments allow reaching a level of sensitivity at the single-cell level: single-cell proteomics, with the use of mass cytometry instruments.
This is why Labtoo developed a tailor-made service to support Labs, Biotechs, Medtech, and Pharma companies in their genomics service outsourcing.
Find the right service for your project: Detection, purification, identification, labeling: all services in proteomics can be found on Labtoo.
Select your Detection of Proteins and Antibodies Service
Conventional techniques for detecting specific proteins in a sample are based on immunodetection, i.e. the detection of an antibody's affinity for its antigen. Thus, an assay using an antibody specific to a protein will indicate the presence or absence of that protein. This is the case, for example, with ELISA or Western blot techniques.

Select your Identification of Peptides and Proteins Service
There is a wide variety of proteins that are constantly being synthesized, modified, and degraded. This diversity contributes to the complex mechanisms that take place both in intra- and extracellular compartments. Identifying the proteins present in a sample is therefore one of the keys to understanding how cells function.
Select your Antibody and Protein Labeling and engineering Service
The conjugation of proteins or peptides with markers is very useful for detection as well as for the study of structure and interactions with other molecules. For example, markers can be fluorescent for imaging analysis or can be used to produce drug-antibody conjugates (DACs).
Select your Protein quantification Service
To determine the amount of a given protein in a sample, immunochemical techniques can be used, such as ELISA or Western Blot. Analytical techniques such as quantitative mass spectrometry or microarray (measurement of interaction with other molecules such as peptides) are also possible.

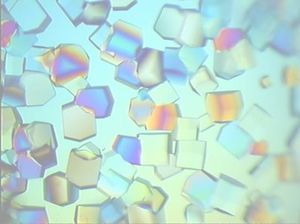
Select your Protein Structure Determination Service
The three-dimensional structure of a protein is directly related to its function, which is why structural analyses of proteins are important. One of the methods commonly used to determine the three-dimensional structure of proteins is the X-ray analysis of protein crystals.
Our team will handle your R&D service management from the beginning to the end
Perform a feasibility study by looking for existing expertise available within the network of partners
Set up a study protocol, financial quotation and preparing contracts with lab partners
Implement the study plan into a schedule, collect all needed materials and reagents and execute the service
Do you need more information regarding services and testing in Proteomics?
Cutting-edge proteomics technologies
Single cell proteomics
While single cell techniques in genomics are widely used, methods for single cell analysis in proteomics are still being developed. How the expressed proteome differs from cell to cell is a critical question, which complements the question addressed by genomics. Mass spectrometry - the state-of-the-art technique in proteomics - can detect and quantify nearly complete and quantified proteomes, starting from tens of thousands of cells or more. Conversely, the technique of mass cytometry makes it possible to quantify proteins, but only in small numbers and on proteins that are sufficiently abundant.
In a typical mammalian cells, reliable analysis by mass spectrometry was only possible for the most abundant proteins. New approaches may soon bring about changes. Nanopore sensors, a now well-established technology for sequencing DNA and RNA at the level of individual cells, are also being investigated for protein sequencing.
Immuno (antibody detection) approaches for protein profiling in single cells are also increasingly being applied on a larger scale.
Targeted proteomics
Targeted proteomics is a general method of detecting a set of proteins of interest. However, the few years of practice have given way to great disparities in approaches, notably by combining several instruments (quadrupoles, orbitrop, MALDI-ToF). Several studies have yielded very good results.
The four methods as classified by Eva Borràs et Eduard Sabidò are the following:
- Selected ion monitoring (SIM): this involves filtering the spectrometer detector for a single peptide, which is used for quantification. The quantification is done in detection per second and can be done sequentially on several peptides.
- Targeted analysis at the peptide level (MS1): The data acquisition is done for all the peptides present in solution over the total duration of the color gradient, which creates an MS1-Map (MS1-Map).
- Targeted analysis at ion fragment level (MS2): One of the major strategies of targeted proteomics, the Selected reaction monitoring (SRM) uses the triple quadrupole MS with a filter on the peptides to be monitored, which will then be analyzed by another MS analyzer. This strategy has a relatively low background noise and high sensitivity.
- Parallel reaction monitoring (PRM): simultaneous analysis of several peptide fragments pre-selected by a quadrupole to isolate the peptide of interest, then a MS in screening mode.
Discovery proteomics
Discovery proteomics is the study and discovery of markers for a given test by mass spectrometry. The number of samples initially tested is relatively small. Once these markers are identified, they are used on a larger scale on a larger number of samples. Advances in discovery are mainly related to the improvement of analytical instruments and software.
Types of providers
Two major categories of proteomics providers:
Academic platforms
Academic platforms are solicited for proteomics requiring the use of expensive equipment or requiring method development. Mass spectrometers are very difficult to finance, which academic structures can do by grouping together. The service is provided by a technological platform available from external structures such as research institutes and companies.
The service companies
Proteomics service companies offer services adapted to the demands of private or public clients: identification, quantification, method development for clinical tests. They can be highly specialized or generalist and can validate methods using GLP (good laboratory practice) principles.
Clinical and diagnostic applications of proteomics
The application of protein (or peptide) biomarkers in clinical studies is a dynamic and rapidly growing field. The introduction of clinical proteomics, such as mass spectrometry-based assays and antibody-based multiplexed protein arrays, has reshaped the landscape of biomarker identification and validation, enabling the discovery of new biomarkers at an unprecedented rate and with unprecedented reliability.
The methods used are typically quantitative mass spectrometry (label-free or SILAC / iTRAQ), capillary electrophoresis coupled to MS (ESI-MS),
The research fields in which proteomic analyses are in progress are the following ones:
Proteomic Analysis in Oncology
Many markers are used in oncology, as is the case for an sponsored by the Montpellier Cancer Institute, which measures five proteins present in the blood (adenylate kinase 2 (AKongoing clinical study2), isocitrate dehydrogenase 2 (IDH2), annexin 1 (ANX1), DNA-apurinic or apyrimidinic site lyase (APEX1), and heat shock cognate 71 kDa (HSC70) ), and which will allow to stratify patient groups and recommend treatments accordingly.
This example is representative of the use of existing treatments at different doses for different patient subgroups, and holds promise for personalized medicine, which can limit certain side effects and be more cost-effective.
Proteomic analysis in nephrological dysfunction
Markers present in urine can be identified and measured by proteomic techniques, as is the case for patients with type 2 diabetes and at risk of developing kidney disease. In a study conducted by the Steno Diabetes Center Copenhagen which has been finalized, a cocktail of peptides (derived from inflammatory proteins, fibrinogens, apolipoproteins, alpha-1 antitrypsin) is analyzed by EC-MS in the urine of 1777 patients, and a standard treatment vs sprironolactone vs placebo is administered randomly.
Proteomic analysis for inflammatory diseases
A classic protein marker of inflammation, C-reactive protein (CRP), is measured with the aim of stratifying subgroups in a cohort of patients under intensive medical observation for persistent infections.
Proteomic analysis for neurological diseases
The S100B protein is a standard biomarker of brain damage and has the advantage of being detected non-invasively since it is found in the blood after blood-brain barrier dysfunction.
This marker is at the heart of several studies, notably in parallel with the study of other markers such as P and Y neuropeptides, neurofilament triplet protein (NFL), or in the context of head trauma follow-up in comparison with head CT scans.
Proteomic analysis for cardiovascular disease
In the case of cardiovascular diseases, a precise and rapid diagnosis is essential. Several ongoing studies are using proteomics to identify different markers, such as the Fatty Acid Binding Protein (FABP), or like this other study that looks at 16 different cardio markers at once (apolipoprotein B100, FABP3, L-selectin, atrionatri-uretic peptide receptor 1, insulin-like growth factor-binding protein 3, coagulation factor V [F5], F9, F10, adiponutrin, von Wille-brand factor, thrombospondin 1, prolactin, paraoxonase 3, epidermal growth factor receptor, vascular endothelial growth factor, henopexin, myeloblastin, plasma serine protease inhibitor, heparin cofactor II, hyaluronan-binding protein).
Used technologies
Protein extraction
Electrophoresis and Western Blot
ELISA and Multiplex ELISA methods
Immunoprecipitation and co-immunoprecipitation
Mass spectrometry
Peptide array
Crystallography and X-ray diffraction

